Research
Research areas of The Gadhave Lab
Overview
The Gadhave Lab studies insect-vectored plant diseases as dynamic systems shaped by biological interactions, environmental variability, and management practices. We integrate insect vector biology, plant virology, and molecular approaches to understand cross-talk among vectors, viruses, and host plants—and how these processes can be predicted and sustainably disrupted.
Our work generates mechanistic insight that scales, linking molecular processes to population-level dynamics and actionable management outcomes.
We organize our research around a unified objective: to build predictive and mechanistic frameworks for the sustainable control of insect-borne plant diseases across scales.
Research Framework
Molecular Pathosystems
Mechanisms governing virus–vector–plant interactions
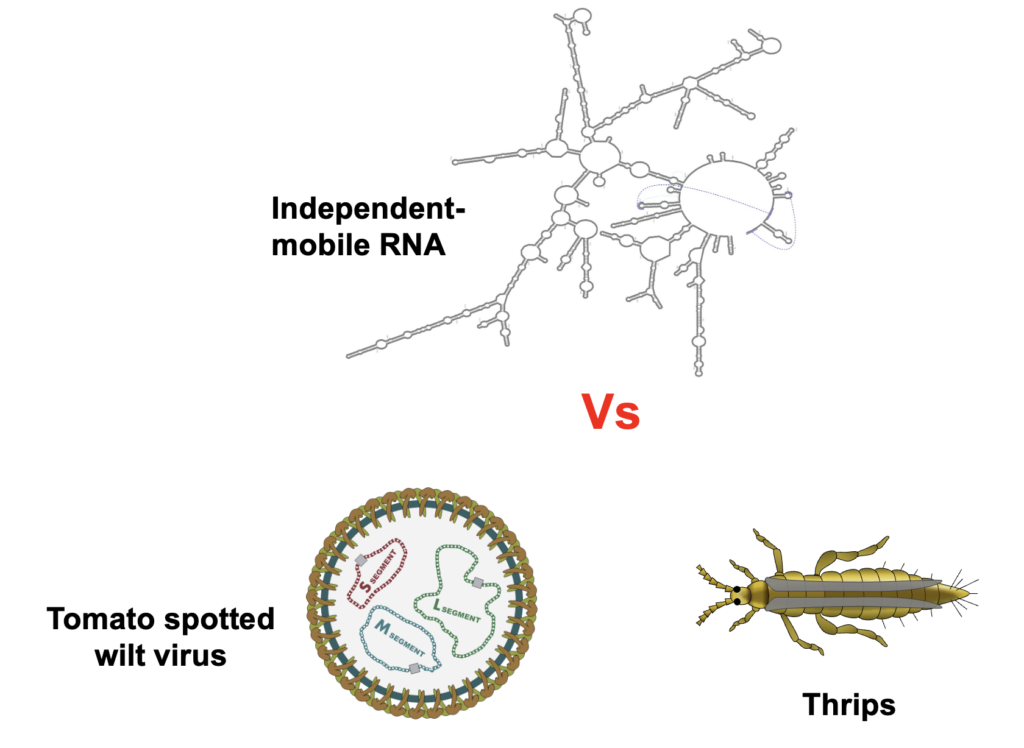
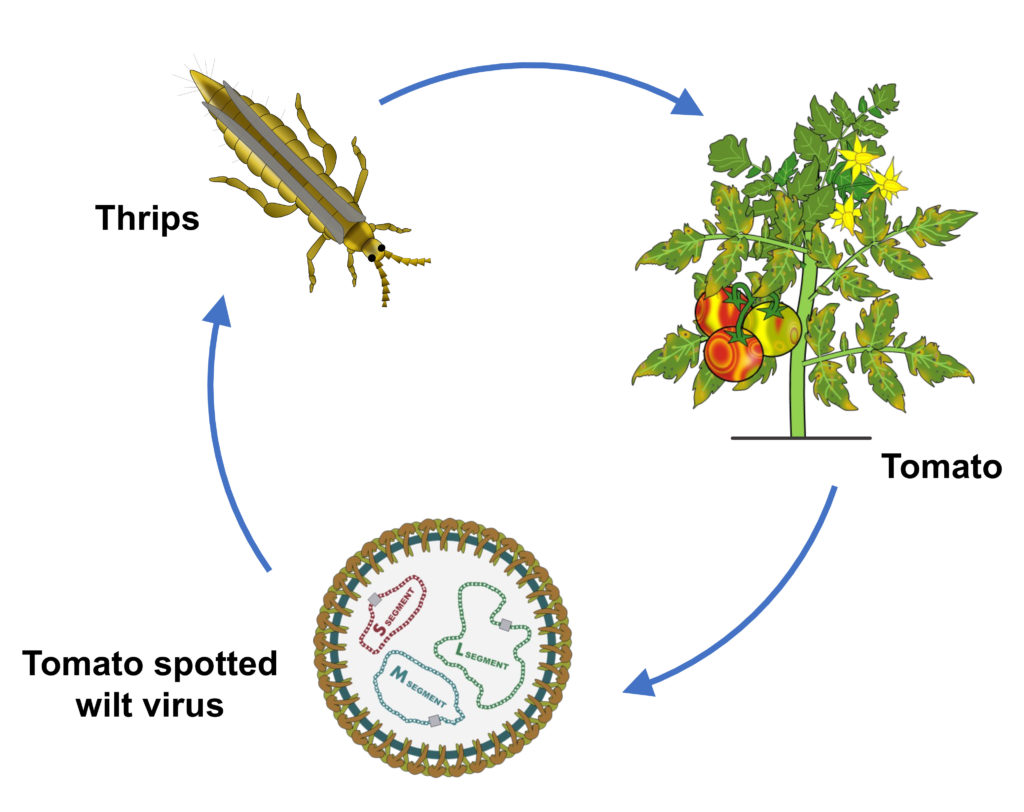
At the core of vector-borne disease are molecular interactions among insects, pathogens, and host plants. We investigate the biological processes that regulate viral pathogenesis, transmission, and host immune evasion, establishing mechanistic foundations for predictive pest control.

Molecular regulation of virus transmission
Mechanistic studies dissect how viruses move within and between vector and plant hosts.
Host plant resistance
We examine how plants respond to viral infection and transmission at molecular and physiological levels.
Determinants of vector competence
We identify molecular factors that influence an insect’s ability to transmit plant viruses.
Scaling molecular processes to epidemiological outcomes
Molecular findings are linked to population-level transmission dynamics and outbreak potential.
RNAi-Based Pest Management
Targeted, insecticide-reducing control strategies
We investigate RNA interference (RNAi) as a precision tool for suppressing insect vectors and viruses with high efficacy. Our work focuses on identifying biologically essential gene targets and evaluating RNAi-based strategies within realistic agricultural contexts.
Identification and validation of RNAi targets
Candidate genes are experimentally validated to determine their effectiveness and specificity in reducing pest and virus performance.
Functional genomics of insect vectors
We identify genes critical for fitness and pathogen transmission using molecular and genomic approaches.
Delivery, stability, and efficacy of RNAi constructs in agricultural systems
We evaluate how RNAi constructs persist and function under controlled environments.
Integration of RNAi into sustainable pest management frameworks
RNAi strategies are assessed as components of broader IPM systems rather than standalone interventions.
Integrated Surveillance & Phenotyping
Scalable measurement of pest and pathogen dynamics
We develop integrated pathogen detection, surveillance and phenotyping pipelines that combine field monitoring, molecular diagnostics, and quantitative analytics to capture pest and pathogen dynamics across spatial and temporal scales.
Field-based monitoring and trapping systems
We design and deploy structured sampling networks to quantify vector populations and disease incidence in real-world settings.
Imaging and machine learning methods for pattern detection
Computational models identify environmental drivers and early warning signals embedded in complex datasets.
CRISPR-enabled diagnostics for rapid pathogen detection
Rapid molecular assays are developed to detect viruses in vectors and plants with high sensitivity and specificity.
Spectral, imaging and sensor-based phenotyping
We use optical and sensor technologies to detect plant stress and disease signatures at fine resolution.
Ecological Intervention & Integrated Pest Management
System-level strategies for sustainable vector control
We translate mechanistic and surveillance insights into ecological and integrated pest management strategies that operate at system scale. Our work examines how environmental variability, agronomic practices, and biological interactions shape vector populations and disease risk.

Landscape and agronomic effects on transmission risk
Field-scale studies evaluate how management practices alter pest & disease pressure.
Environmental drivers of vector population dynamics
We quantify how climate, cropping systems, and habitat structure influence vector abundance and movement.
Ecologically informed intervention strategies
Intervention approaches are designed to exploit biological vulnerabilities rather than rely solely on chemical suppression.
Integration of predictive tools into IPM frameworks
Forecasting and diagnostic tools are incorporated into decision-support systems to enable proactive management.

Cross-Cutting Platforms
Across all pillars, the lab employs shared analytical and experimental platforms that enable integration and scalability:
- Long-term, multi-site field datasets: These datasets provide ecological context and support longitudinal analyses of vector and disease dynamics.
- Molecular diagnostics and sensing technologies: Advanced detection tools allow rapid and precise quantification of pathogens in vectors and plants.
- Quantitative modeling and machine learning approaches: Computational frameworks integrate heterogeneous data to identify drivers and forecast outcomes.
- Integrated experimental–computational workflows: Iterative feedback between laboratory experiments and field data strengthens predictive accuracy.
